Introduction
Let’s understand Machine Learning Distances through a simple example.
Imagine planning a vacation to a city with an attraction you want to visit. To avoid wasting time traveling, you search for a hotel near this attraction.
But how can you evaluate the distances between several potential hotels to find the closest one?
This typical real-life situation translates well to data analysis. Instead of measuring the distance between hotels and attractions, we want to determine if data points—for example, from two different patients—are similar (close) or dissimilar (far apart).
Many machine learning algorithms are built on methods developed to solve this fundamental question.
Imagine our dataset as a set of points in space, where each observation (or record) represents a single point.
For example, a patient (x) can be described by several features: blood pressure (x₁), cholesterol (x₂), body mass index (x₃), and blood glucose (x₄).
Since we have four features, each patient can be represented by a point in a four-dimensional space.
| Patient | Pressure (mmHg) | Cholesterol (mg/dL) | BMI | Blood glucose (mg/dL) |
| A | 140 | 220 | 27 | 100 |
| B | 130 | 190 | 25 | 90 |
| C | 160 | 250 | 30 | 120 |
The points can be summarized as follows:
A = (140, 220, 27, 100)
B = (130, 190, 25, 90)
C = (160, 250, 30, 120)
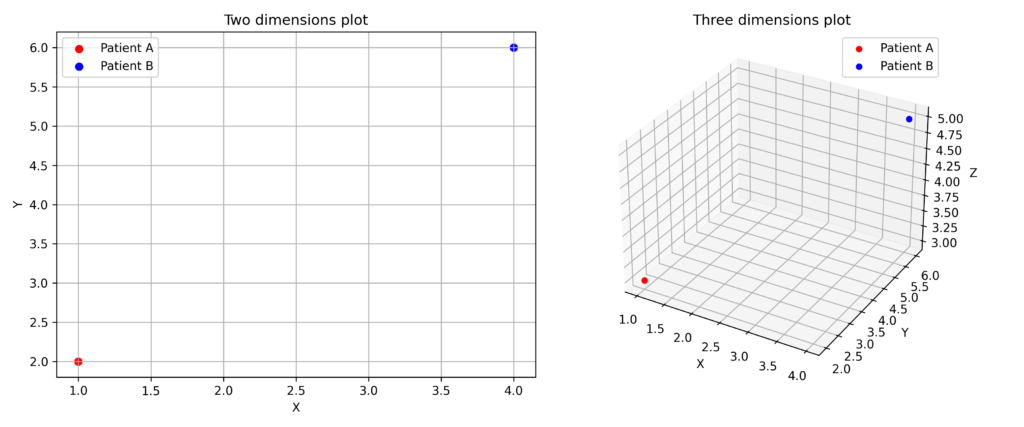
This mathematical space containing these points is called the feature space, where each point is represented as a vector. Depending on the number of features used, the space can have any number of dimensions.
![]()
Visualizing the feature space is straightforward in two dimensions, more challenging in three dimensions, and increasingly difficult in higher-dimensional spaces—where our visual capabilities fall short.
Distance Measurements
Machine learning offers numerous methods for calculating distances between points:
Euclidean Distance
Mahnattan Distance
Minkowski Distance
Hamming Distance
Cosine Distance
Mahalanobis Distance
Jaccard Distance
Bray-Curtis Distance
Hellinger Distance
Let’s explore the most widely used approaches.
Euclidean Distance
Euclidean distance is the simplest way to measure how far apart two points are. In a two-dimensional plane, it’s the length of the straight line that connects them.
This distance measure comes from the Pythagorean theorem, which tells us that in a right triangle, the square of the hypotenuse equals the sum of squares of the other two sides. For two points A and B with coordinates x₁y₁ and x₂y₂, we can apply this theorem:
![]()
We can extend this to calculate the distance between any two points in an n-dimensional space:
![]()
To see this in action, let’s look at two patients, A and B. Patient A has a blood pressure of 140 mmHg and cholesterol of 250 mg/dl, while Patient B has values of 130 mmHg and 220 mg/dl respectively.
Here’s how we calculate their Euclidean distance:
![]()
![]()
![]()
![]()
This value of 31.62 tells us how different these patients are: a smaller number means the patients are more similar.
Euclidean distance works best under these conditions:
- The features are continuous numbers with similar scales
- The dataset has relatively few dimensions. With too many dimensions, Euclidean distance loses effectiveness due to the Curse of Dimensionality—as dimensions increase, the distances between points become more uniform and less meaningful.
Manhattan Distance
Manhattan distance (also known as “city block distance” or “grid distance”) measures the path between points using only vertical and horizontal movements, not diagonal ones. The name comes from Manhattan’s street grid layout in New York City, where streets intersect at right angles.
The Manhattan distance formula is:
![]()
This method simply adds up the absolute differences between corresponding points, without squaring them like in Euclidean distance.
Manhattan distance is particularly effective when working with data that naturally follows a grid pattern—such as medical images—or when analyzing categorical data represented by integers.
Minkowski Distance
The Minkowski distance generalizes distance calculations between points into a single formula that can function as either Euclidean or Manhattan distance, depending on the parameter p.
When p equals 1, Minkowski distance acts as Manhattan distance; when p equals 2, it becomes Euclidean distance. Values of p greater than 2 place more emphasis on larger differences between measurements.
The Minkowski distance formula is:
![]()
By adjusting the parameter p, the Minkowski distance lets us evaluate distances using different methods—making it particularly valuable when we’re uncertain which method will work best.
Hamming Distance
For non-numerical data (like blood types or medications), traditional distance calculations aren’t suitable. The Hamming method offers a solution by counting the differences between features across observations.
Consider two patients’ therapies: Patient A takes Aspirin and Insulin, while Patient B takes Paracetamol and Insulin.
Using the Hamming method, the distance between these patients is 1—representing their single difference in medication.
The Hamming distance formula is as follows:
![]()
This formula effectively compares categorical data or strings (such as DNA sequences).
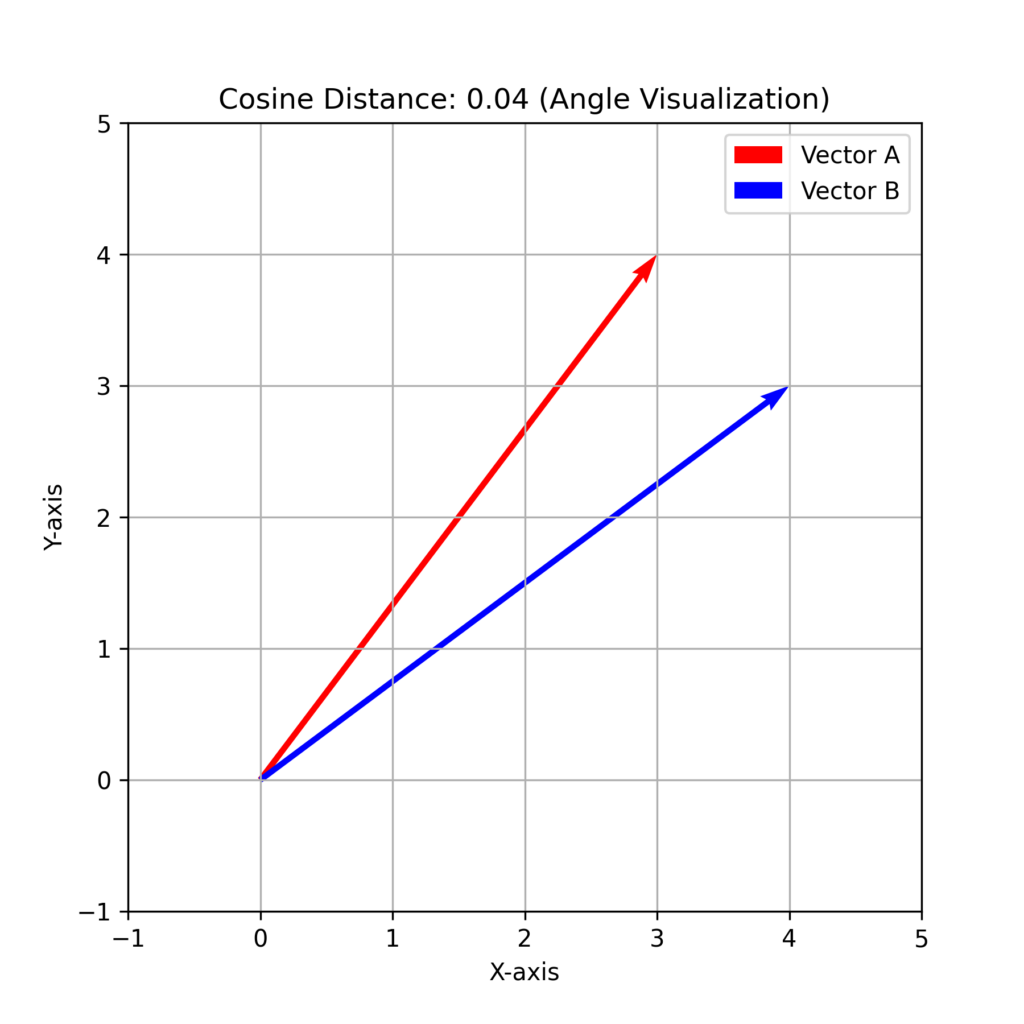
Cosine Distance
Cosine distance measures the angle between two vectors rather than their absolute difference (magnitude). Since it uses cosine, the result ranges from 0 (identical vectors) to 1 (completely different vectors).
![]()
This distance measure is particularly useful for vector-based features, such as text documents or image data.
Mahalanobis Distance
The Mahalanobis distance measures how far apart points are while accounting for both the correlation between variables and their variance.
![]()
Here, S⁻¹ represents the covariance matrix of the dataset.
This distance measure is particularly useful for datasets with correlated features (such as blood glucose and insulin levels). When features are uncorrelated, it yields results similar to Euclidean distance.
Jaccard Distance
The Jaccard distance measures the dissimilarity between two sets by comparing their shared and unique elements.
![]()
This measure yields values between 0 and 1, where 0 indicates identical sets and 1 indicates completely distinct sets.
In medicine, the Jaccard distance helps analyze symptom patterns for disease prediction and compare DNA sequences.
Bray-Curtis Distance
The Bray-Curtis distance measures the relative difference between two vectors.
![]()
This metric is particularly useful for comparing proportions, such as the ratio between lean mass and body fat, or analyzing differences in bacterial population compositions across samples.
Hellinger Distance
This metric compares probability distributions to measure their similarity.
![]()
It helps analyze differences in marker distributions between healthy and sick subjects and detect bias in medical data.
Summary table
| Metric | Data Type | Advantages | Disadvantages |
|---|---|---|---|
| Euclidean | Continuous numbers | Intuitive to understand | Sensitive to scale differences |
| Manhattan | Grid-based data | Robust against outliers | Less accurate with continuous data |
| Minkowski | Generalizes Euclidean and Manhattan | Highly flexible | Requires parameter p selection |
| Cosine | Text, images | Magnitude-independent | Poor performance with zero values |
| Mahalanobis | Correlated data | Accounts for variance | Requires covariance computation |
| Jaccard | Sets, binary data | Works well with categories | Ignores difference magnitudes |
| Bray-Curtis | Compositional data | Handles proportions well | Cannot process negative values |
| Hellinger | Probability distributions | Ideal for probabilistic data | Needs distribution-formatted data |
Python Implementation of Distance Metrics
The script creates a synthetic dataset with medical features (Systolic Pressure, Cholesterol, Glucose, and BMI). It then selects two random patients and calculates various distances between them using Euclidean, Manhattan, Minkowski (p=3), Cosine, Mahalanobis, Jaccard, Bray-Curtis, Hellinger, and Hamming metrics.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
def euclidean_distance(x, y):
return np.sqrt(np.sum((x - y) ** 2))
def manhattan_distance(x, y):
return np.sum(np.abs(x - y))
def minkowski_distance(x, y, p):
return np.sum(np.abs(x - y) ** p) ** (1 / p)
def cosine_distance(x, y):
return 1 - (np.dot(x, y) / (np.linalg.norm(x) * np.linalg.norm(y)))
def mahalanobis_distance(x, y, cov_inv):
diff = x - y
return np.sqrt(np.dot(np.dot(diff.T, cov_inv), diff))
def jaccard_distance(x, y):
intersection = np.sum(np.minimum(x, y))
union = np.sum(np.maximum(x, y))
return 1 - intersection / union
def bray_curtis_distance(x, y):
return np.sum(np.abs(x - y)) / np.sum(x + y)
def hellinger_distance(x, y):
x = np.array(x, dtype=np.float64)
y = np.array(y, dtype=np.float64)
return (1 / np.sqrt(2)) * np.sqrt(np.sum((np.sqrt(x + 1e-10) - np.sqrt(y + 1e-10)) ** 2))
# Generate a synthetic dataset with medical data
np.random.seed(42)
num_patients = 10
data = {
"Patient": [f"P{i+1}" for i in range(num_patients)],
"Systolic_Pressure": np.random.randint(100, 180, num_patients), # mmHg
"Cholesterol": np.random.randint(150, 250, num_patients), # mg/dL
"Glucose": np.random.randint(70, 200, num_patients), # mg/dL
"BMI": np.round(np.random.uniform(18, 35, num_patients), 1), # kg/m^2
"Smoking": np.random.choice(["Yes", "No"], num_patients), # Categorical
}
df = pd.DataFrame(data)
# Normalize numerical features to avoid scale imbalances
scaler = MinMaxScaler()
df_scaled = df.copy()
df_scaled.iloc[:, 1:-1] = scaler.fit_transform(df.iloc[:, 1:-1])
# Convert categorical column "Smoking" into numerical for Hamming distance
df_scaled["Smoking"] = df["Smoking"].map({"No": 0, "Yes": 1})
# Compute covariance matrix for Mahalanobis distance
cov_matrix = np.cov(df_scaled.iloc[:, 1:-1].T)
cov_inv = np.linalg.inv(cov_matrix)
# Select two random patients to compute distances
p1, p2 = np.random.choice(df_scaled.index, 2, replace=False)
X_p1 = df_scaled.iloc[p1, 1:].values
X_p2 = df_scaled.iloc[p2, 1:].values
# Compute distances
dist_euclidean = euclidean_distance(X_p1, X_p2)
dist_manhattan = manhattan_distance(X_p1, X_p2)
dist_minkowski_3 = minkowski_distance(X_p1, X_p2, 3)
dist_cosine = cosine_distance(X_p1, X_p2)
dist_mahalanobis = mahalanobis_distance(X_p1[:-1], X_p2[:-1], cov_inv)
dist_jaccard = jaccard_distance(X_p1[:-1], X_p2[:-1])
dist_bray_curtis = bray_curtis_distance(X_p1[:-1], X_p2[:-1])
dist_hellinger = hellinger_distance(X_p1[:-1], X_p2[:-1])
dist_hamming = np.sum(X_p1[:-1] != X_p2[:-1]) # Excluding categorical variable
# Create a results dataframe
dist_results = pd.DataFrame({
"Distance": ["Euclidean", "Manhattan", "Minkowski (p=3)", "Cosine", "Mahalanobis", "Jaccard", "Bray-Curtis", "Hellinger", "Hamming"],
"Value": [dist_euclidean, dist_manhattan, dist_minkowski_3, dist_cosine, dist_mahalanobis, dist_jaccard, dist_bray_curtis, dist_hellinger, dist_hamming]
})
# Plot the distance comparison
fig, ax = plt.subplots(figsize=(10, 6))
sns.barplot(x=dist_results["Distance"], y=dist_results["Value"], ax=ax, palette="coolwarm")
ax.set_title("Comparison of Different Distance Metrics", fontsize=14)
ax.set_ylabel("Distance Value", fontsize=12)
ax.set_xlabel("Distance Type", fontsize=12)
ax.grid(axis="y", linestyle="--", alpha=0.7)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45, ha="right")
# Annotate bars with values
for i, v in enumerate(dist_results["Value"]):
ax.text(i, v + 0.05, f"{v:.2f}", ha='center', fontsize=12)
plt.show()
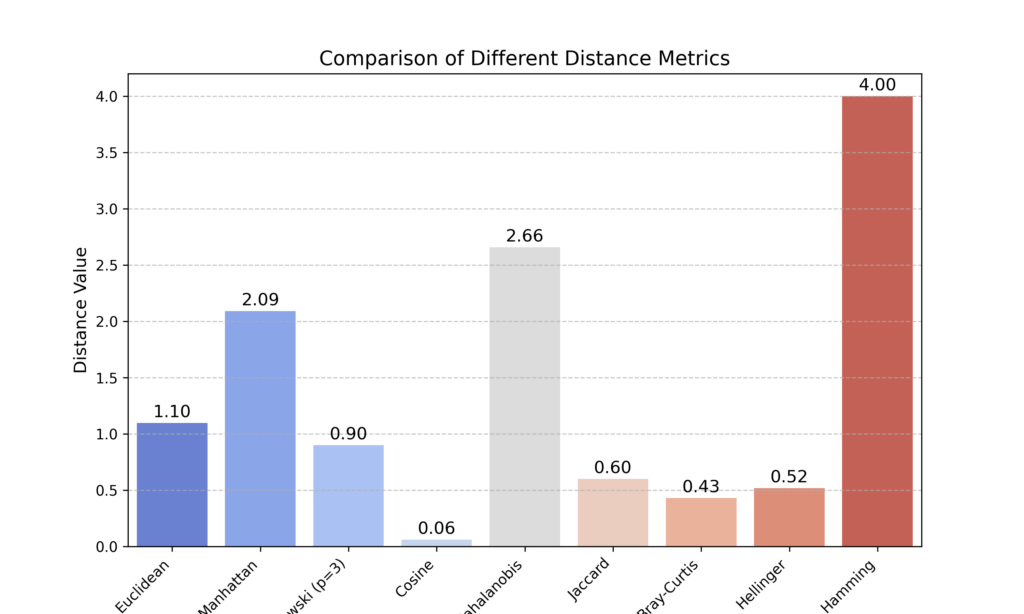
While the results are plotted to compare these distance metrics, it’s important to note that each metric serves a specific purpose based on the dataset’s characteristics.
However, the resulting graph is potentially misleading since each metric uses different formulas and has distinct interpretations that must be considered in context.
For example, the Cosine, Jaccard, Bray-Curtis, and Hellinger methods have values that range between 0 and 1, while other methods can have any value.
Additional Resources
Juan Luis Suárez, Salvador García, Francisco Herrera, A tutorial on distance metric learning: Mathematical foundations, algorithms, experimental analysis, prospects and challenges, Neurocomputing, Volume 425, 2021, Pages 300-322, ISSN 0925-2312
Kaggle. Distance measures in machine learning
Conclusions
Selecting the right distance metric is crucial for successful machine learning analysis. Rather than having one “best” metric, different metrics suit different types of data and goals:
- Euclidean distance works best for continuous numerical data in low-dimensional spaces
- Manhattan distance suits data organized in grid-like structures
- Mahalanobis distance excels when working with correlated variables
- Hamming or Jaccard distances are ideal for categorical or binary data
Keep in mind the computational costs: some metrics, such as Mahalanobis, involve complex calculations that may be impractical for large datasets.
Before choosing a distance metric, consider these key factors:
- The type of data you’re working with (numerical, categorical, or mixed)
- Whether your variables are correlated
- The number of dimensions in your feature space
- Whether your data needs normalization to ensure fair comparisons
In practice, it’s often valuable to experiment with different distance metrics and determine which one yields the most meaningful results for your specific problem.


